numpy.random.Generator.multivariate_normal#
method
- random.Generator.multivariate_normal(mean, cov, size=None, check_valid='warn', tol=1e-8, *, method='svd')#
Draw random samples from a multivariate normal distribution.
The multivariate normal, multinormal or Gaussian distribution is a generalization of the one-dimensional normal distribution to higher dimensions. Such a distribution is specified by its mean and covariance matrix. These parameters are analogous to the mean (average or “center”) and variance (standard deviation, or “width,” squared) of the one-dimensional normal distribution.
- Parameters:
- mean1-D array_like, of length N
Mean of the N-dimensional distribution.
- cov2-D array_like, of shape (N, N)
Covariance matrix of the distribution. It must be symmetric and positive-semidefinite for proper sampling.
- sizeint or tuple of ints, optional
Given a shape of, for example,
(m,n,k),m*n*ksamples are generated, and packed in an m-by-n-by-k arrangement. Because each sample is N-dimensional, the output shape is(m,n,k,N). If no shape is specified, a single (N-D) sample is returned.- check_valid{ ‘warn’, ‘raise’, ‘ignore’ }, optional
Behavior when the covariance matrix is not positive semidefinite.
- tolfloat, optional
Tolerance when checking the singular values in covariance matrix. cov is cast to double before the check.
- method{ ‘svd’, ‘eigh’, ‘cholesky’}, optional
The cov input is used to compute a factor matrix A such that
A @ A.T = cov. This argument is used to select the method used to compute the factor matrix A. The default method ‘svd’ is the slowest, while ‘cholesky’ is the fastest but less robust than the slowest method. The method eigh uses eigen decomposition to compute A and is faster than svd but slower than cholesky.New in version 1.18.0.
- Returns:
- outndarray
The drawn samples, of shape size, if that was provided. If not, the shape is
(N,).In other words, each entry
out[i,j,...,:]is an N-dimensional value drawn from the distribution.
Notes
The mean is a coordinate in N-dimensional space, which represents the location where samples are most likely to be generated. This is analogous to the peak of the bell curve for the one-dimensional or univariate normal distribution.
Covariance indicates the level to which two variables vary together. From the multivariate normal distribution, we draw N-dimensional samples, \(X = [x_1, x_2, ... x_N]\). The covariance matrix element \(C_{ij}\) is the covariance of \(x_i\) and \(x_j\). The element \(C_{ii}\) is the variance of \(x_i\) (i.e. its “spread”).
Instead of specifying the full covariance matrix, popular approximations include:
This geometrical property can be seen in two dimensions by plotting generated data-points:
>>> mean = [0, 0] >>> cov = [[1, 0], [0, 100]] # diagonal covariance
Diagonal covariance means that points are oriented along x or y-axis:
>>> import matplotlib.pyplot as plt >>> x, y = np.random.default_rng().multivariate_normal(mean, cov, 5000).T >>> plt.plot(x, y, 'x') >>> plt.axis('equal') >>> plt.show()
Note that the covariance matrix must be positive semidefinite (a.k.a. nonnegative-definite). Otherwise, the behavior of this method is undefined and backwards compatibility is not guaranteed.
References
[1]Papoulis, A., “Probability, Random Variables, and Stochastic Processes,” 3rd ed., New York: McGraw-Hill, 1991.
[2]Duda, R. O., Hart, P. E., and Stork, D. G., “Pattern Classification,” 2nd ed., New York: Wiley, 2001.
Examples
>>> mean = (1, 2) >>> cov = [[1, 0], [0, 1]] >>> rng = np.random.default_rng() >>> x = rng.multivariate_normal(mean, cov, (3, 3)) >>> x.shape (3, 3, 2)
We can use a different method other than the default to factorize cov:
>>> y = rng.multivariate_normal(mean, cov, (3, 3), method='cholesky') >>> y.shape (3, 3, 2)
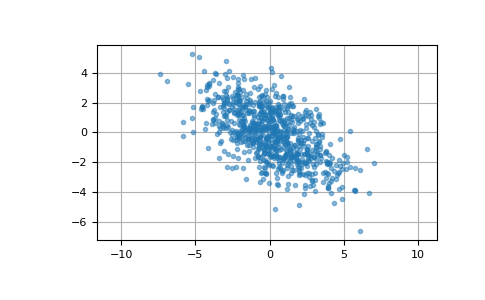
Here we generate 800 samples from the bivariate normal distribution with mean [0, 0] and covariance matrix [[6, -3], [-3, 3.5]]. The expected variances of the first and second components of the sample are 6 and 3.5, respectively, and the expected correlation coefficient is -3/sqrt(6*3.5) ≈ -0.65465.
>>> cov = np.array([[6, -3], [-3, 3.5]]) >>> pts = rng.multivariate_normal([0, 0], cov, size=800)
Check that the mean, covariance, and correlation coefficient of the sample are close to the expected values:
>>> pts.mean(axis=0) array([ 0.0326911 , -0.01280782]) # may vary >>> np.cov(pts.T) array([[ 5.96202397, -2.85602287], [-2.85602287, 3.47613949]]) # may vary >>> np.corrcoef(pts.T)[0, 1] -0.6273591314603949 # may vary
We can visualize this data with a scatter plot. The orientation of the point cloud illustrates the negative correlation of the components of this sample.
>>> import matplotlib.pyplot as plt >>> plt.plot(pts[:, 0], pts[:, 1], '.', alpha=0.5) >>> plt.axis('equal') >>> plt.grid() >>> plt.show()