Determining Moore’s Law with real data in NumPy#
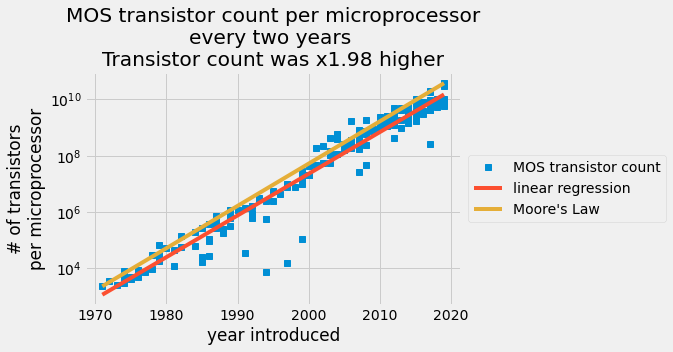
The number of transistors reported per a given chip plotted on a log scale in the y axis with the date of introduction on the linear scale x-axis. The blue data points are from a transistor count table. The red line is an ordinary least squares prediction and the orange line is Moore’s law.
What you’ll do#
In 1965, engineer Gordon Moore predicted that transistors on a chip would double every two years in the coming decade [1]. You’ll compare Moore’s prediction against actual transistor counts in the 53 years following his prediction. You will determine the best-fit constants to describe the exponential growth of transistors on semiconductors compared to Moore’s Law.
Skills you’ll learn#
Load data from a *.csv file
Perform linear regression and predict exponential growth using ordinary least squares
You’ll compare exponential growth constants between models
Share your analysis in a file:
as NumPy zipped files
*.npzas a
*.csvfile
Assess the amazing progress semiconductor manufacturers have made in the last five decades
What you’ll need#
1. These packages:
NumPy
imported with the following commands
import matplotlib.pyplot as plt
import numpy as np
2. Since this is an exponential growth law you need a little background in doing math with natural logs and exponentials.
You’ll use these NumPy and Matplotlib functions:
np.loadtxt: this function loads text into a NumPy arraynp.log: this function takes the natural log of all elements in a NumPy arraynp.exp: this function takes the exponential of all elements in a NumPy arraylambda: this is a minimal function definition for creating a function modelplt.semilogy: this function will plot x-y data onto a figure with a linear x-axis and \(\log_{10}\) y-axisplt.plot: this function will plot x-y data on linear axesslicing arrays: view parts of the data loaded into the workspace, slice the arrays e.g.
x[:10]for the first 10 values in the array,xboolean array indexing: to view parts of the data that match a given condition use boolean operations to index an array
np.block: to combine arrays into 2D arraysnp.newaxis: to change a 1D vector to a row or column vectornp.savezandnp.savetxt: these two functions will save your arrays in zipped array format and text, respectively
Building Moore’s law as an exponential function#
Your empirical model assumes that the number of transistors per semiconductor follows an exponential growth,
\(\log(\text{transistor_count})= f(\text{year}) = A\cdot \text{year}+B,\)
where \(A\) and \(B\) are fitting constants. You use semiconductor manufacturers’ data to find the fitting constants.
You determine these constants for Moore’s law by specifying the rate for added transistors, 2, and giving an initial number of transistors for a given year.
You state Moore’s law in an exponential form as follows,
\(\text{transistor_count}= e^{A_M\cdot \text{year} +B_M}.\)
Where \(A_M\) and \(B_M\) are constants that double the number of transistors every two years and start at 2250 transistors in 1971,
\(\dfrac{\text{transistor_count}(\text{year} +2)}{\text{transistor_count}(\text{year})} = 2 = \dfrac{e^{B_M}e^{A_M \text{year} + 2A_M}}{e^{B_M}e^{A_M \text{year}}} = e^{2A_M} \rightarrow A_M = \frac{\log(2)}{2}\)
\(\log(2250) = \frac{\log(2)}{2}\cdot 1971 + B_M \rightarrow B_M = \log(2250)-\frac{\log(2)}{2}\cdot 1971\)
so Moore’s law stated as an exponential function is
\(\log(\text{transistor_count})= A_M\cdot \text{year}+B_M,\)
where
\(A_M=0.3466\)
\(B_M=-675.4\)
Since the function represents Moore’s law, define it as a Python
function using
lambda
A_M = np.log(2) / 2
B_M = np.log(2250) - A_M * 1971
Moores_law = lambda year: np.exp(B_M) * np.exp(A_M * year)
In 1971, there were 2250 transistors on the Intel 4004 chip. Use
Moores_law to check the number of semiconductors Gordon Moore would expect
in 1973.
ML_1971 = Moores_law(1971)
ML_1973 = Moores_law(1973)
print("In 1973, G. Moore expects {:.0f} transistors on Intels chips".format(ML_1973))
print("This is x{:.2f} more transistors than 1971".format(ML_1973 / ML_1971))
In 1973, G. Moore expects 4500 transistors on Intels chips
This is x2.00 more transistors than 1971
Loading historical manufacturing data to your workspace#
Now, make a prediction based upon the historical data for
semiconductors per chip. The Transistor Count
[3]
each year is in the transistor_data.csv file. Before loading a *.csv
file into a NumPy array, its a good idea to inspect the structure of the
file first. Then, locate the columns of interest and save them to a
variable. Save two columns of the file to the array, data.
Here, print out the first 10 rows of transistor_data.csv. The columns are
Processor |
MOS transistor count |
Date of Introduction |
Designer |
MOSprocess |
Area |
|---|---|---|---|---|---|
Intel 4004 (4-bit 16-pin) |
2250 |
1971 |
Intel |
“10,000 nm” |
12 mm² |
… |
… |
… |
… |
… |
… |
! head transistor_data.csv
Processor,MOS transistor count,Date of Introduction,Designer,MOSprocess,Area
Intel 4004 (4-bit 16-pin),2250,1971,Intel,"10,000 nm",12 mm²
Intel 8008 (8-bit 18-pin),3500,1972,Intel,"10,000 nm",14 mm²
NEC μCOM-4 (4-bit 42-pin),2500,1973,NEC,"7,500 nm",?
Intel 4040 (4-bit 16-pin),3000,1974,Intel,"10,000 nm",12 mm²
Motorola 6800 (8-bit 40-pin),4100,1974,Motorola,"6,000 nm",16 mm²
Intel 8080 (8-bit 40-pin),6000,1974,Intel,"6,000 nm",20 mm²
TMS 1000 (4-bit 28-pin),8000,1974,Texas Instruments,"8,000 nm",11 mm²
MOS Technology 6502 (8-bit 40-pin),4528,1975,MOS Technology,"8,000 nm",21 mm²
Intersil IM6100 (12-bit 40-pin; clone of PDP-8),4000,1975,Intersil,,
You don’t need the columns that specify Processor, Designer, MOSprocess, or Area. That leaves the second and third columns, MOS transistor count and Date of Introduction, respectively.
Next, you load these two columns into a NumPy array using
np.loadtxt.
The extra options below will put the data in the desired format:
delimiter = ',': specify delimeter as a comma ‘,’ (this is the default behavior)usecols = [1,2]: import the second and third columns from the csvskiprows = 1: do not use the first row, because its a header row
data = np.loadtxt("transistor_data.csv", delimiter=",", usecols=[1, 2], skiprows=1)
You loaded the entire history of semiconducting into a NumPy array named
data. The first column is the MOS transistor count and the second
column is the Date of Introduction in a four-digit year.
Next, make the data easier to read and manage by assigning the two
columns to variables, year and transistor_count. Print out the first
10 values by slicing the year and transistor_count arrays with
[:10]. Print these values out to check that you have the saved the
data to the correct variables.
year = data[:, 1] # grab the second column and assign
transistor_count = data[:, 0] # grab the first column and assign
print("year:\t\t", year[:10])
print("trans. cnt:\t", transistor_count[:10])
year: [1971. 1972. 1973. 1974. 1974. 1974. 1974. 1975. 1975. 1975.]
trans. cnt: [2250. 3500. 2500. 3000. 4100. 6000. 8000. 4528. 4000. 5000.]
You are creating a function that predicts the transistor count given a
year. You have an independent variable, year, and a dependent
variable, transistor_count. Transform the dependent variable to
log-scale,
\(y_i = \log(\) transistor_count[i] \(),\)
resulting in a linear equation,
\(y_i = A\cdot \text{year} +B\).
yi = np.log(transistor_count)
Calculating the historical growth curve for transistors#
Your model assume that yi is a function of year. Now, find the best-fit model that minimizes the difference between \(y_i\) and \(A\cdot \text{year} +B, \) as such
\(\min \sum|y_i - (A\cdot \text{year}_i + B)|^2.\)
This sum of squares error can be succinctly represented as arrays as such
\(\sum|\mathbf{y}-\mathbf{Z} [A,~B]^T|^2,\)
where \(\mathbf{y}\) are the observations of the log of the number of transistors in a 1D array and \(\mathbf{Z}=[\text{year}_i^1,~\text{year}_i^0]\) are the polynomial terms for \(\text{year}_i\) in the first and second columns. By creating this set of regressors in the \(\mathbf{Z}-\)matrix you set up an ordinary least squares statistical model.
Z is a linear model with two parameters, i.e. a polynomial with degree 1.
Therefore we can represent the model with numpy.polynomial.Polynomial and
use the fitting functionality to determine the model parameters:
model = np.polynomial.Polynomial.fit(year, yi, deg=1)
By default, Polynomial.fit performs the fit in the domain determined by the
independent variable (year in this case).
The coefficients for the unscaled and unshifted model can be recovered with the
convert method:
model = model.convert()
model
The individual parameters \(A\) and \(B\) are the coefficients of our linear model:
B, A = model
Did manufacturers double the transistor count every two years? You have the final formula,
\(\dfrac{\text{transistor_count}(\text{year} +2)}{\text{transistor_count}(\text{year})} = xFactor = \dfrac{e^{B}e^{A( \text{year} + 2)}}{e^{B}e^{A \text{year}}} = e^{2A}\)
where increase in number of transistors is \(xFactor,\) number of years is 2, and \(A\) is the best fit slope on the semilog function.
print(f"Rate of semiconductors added on a chip every 2 years: {np.exp(2 * A):.2f}")
Rate of semiconductors added on a chip every 2 years: 1.98
Based upon your least-squares regression model, the number of semiconductors per chip increased by a factor of \(1.98\) every two years. You have a model that predicts the number of semiconductors each year. Now compare your model to the actual manufacturing reports. Plot the linear regression results and all of the transistor counts.
Here, use
plt.semilogy
to plot the number of transistors on a log-scale and the year on a
linear scale. You have defined a three arrays to get to a final model
\(y_i = \log(\text{transistor_count}),\)
\(y_i = A \cdot \text{year} + B,\)
and
\(\log(\text{transistor_count}) = A\cdot \text{year} + B,\)
your variables, transistor_count, year, and yi all have the same
dimensions, (179,). NumPy arrays need the same dimensions to make a
plot. The predicted number of transistors is now
\(\text{transistor_count}_{\text{predicted}} = e^Be^{A\cdot \text{year}}\).
In the next plot, use the
fivethirtyeight
style sheet.
The style sheet replicates
https://fivethirtyeight.com elements. Change the matplotlib style with
plt.style.use.
transistor_count_predicted = np.exp(B) * np.exp(A * year)
transistor_Moores_law = Moores_law(year)
plt.style.use("fivethirtyeight")
plt.semilogy(year, transistor_count, "s", label="MOS transistor count")
plt.semilogy(year, transistor_count_predicted, label="linear regression")
plt.plot(year, transistor_Moores_law, label="Moore's Law")
plt.title(
"MOS transistor count per microprocessor\n"
+ "every two years \n"
+ "Transistor count was x{:.2f} higher".format(np.exp(A * 2))
)
plt.xlabel("year introduced")
plt.legend(loc="center left", bbox_to_anchor=(1, 0.5))
plt.ylabel("# of transistors\nper microprocessor")
Text(0, 0.5, '# of transistors\nper microprocessor')
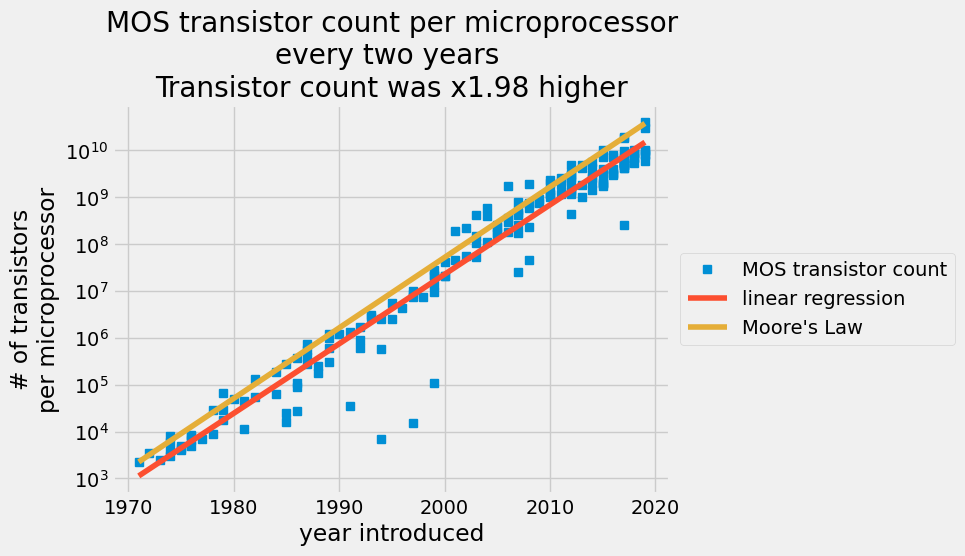
A scatter plot of MOS transistor count per microprocessor every two years with a red line for the ordinary least squares prediction and an orange line for Moore’s law.
The linear regression captures the increase in the number of transistors per semiconductors each year. In 2015, semiconductor manufacturers claimed they could not keep up with Moore’s law anymore. Your analysis shows that since 1971, the average increase in transistor count was x1.98 every 2 years, but Gordon Moore predicted it would be x2 every 2 years. That is an amazing prediction.
Consider the year 2017. Compare the data to your linear regression model and Gordon Moore’s prediction. First, get the transistor counts from the year 2017. You can do this with a Boolean comparator,
year == 2017.
Then, make a prediction for 2017 with Moores_law defined above
and plugging in your best fit constants into your function
\(\text{transistor_count} = e^{B}e^{A\cdot \text{year}}\).
A great way to compare these measurements is to compare your prediction
and Moore’s prediction to the average transistor count and look at the
range of reported values for that year. Use the
plt.plot
option,
alpha=0.2,
to increase the transparency of the data. The more opaque the points
appear, the more reported values lie on that measurement. The green \(+\)
is the average reported transistor count for 2017. Plot your predictions
for $\pm\frac{1}{2}~years.
transistor_count2017 = transistor_count[year == 2017]
print(
transistor_count2017.max(), transistor_count2017.min(), transistor_count2017.mean()
)
y = np.linspace(2016.5, 2017.5)
your_model2017 = np.exp(B) * np.exp(A * y)
Moore_Model2017 = Moores_law(y)
plt.plot(
2017 * np.ones(np.sum(year == 2017)),
transistor_count2017,
"ro",
label="2017",
alpha=0.2,
)
plt.plot(2017, transistor_count2017.mean(), "g+", markersize=20, mew=6)
plt.plot(y, your_model2017, label="Your prediction")
plt.plot(y, Moore_Model2017, label="Moores law")
plt.ylabel("# of transistors\nper microprocessor")
plt.legend()
19200000000.0 250000000.0 7050000000.0
<matplotlib.legend.Legend at 0x7fc7a110fed0>
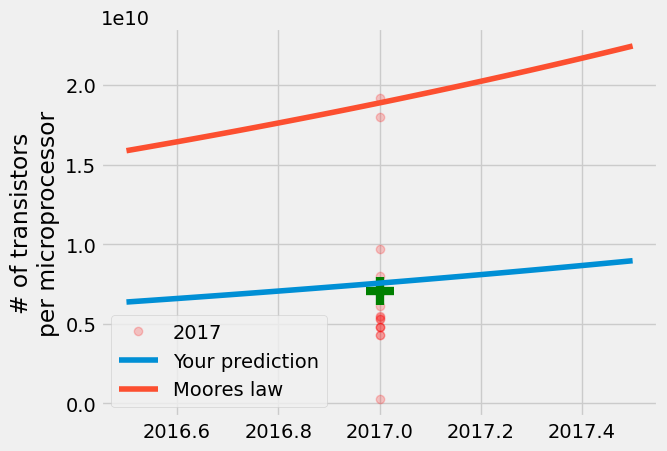
The result is that your model is close to the mean, but Gordon Moore’s prediction is closer to the maximum number of transistors per microprocessor produced in 2017. Even though semiconductor manufacturers thought that the growth would slow, once in 1975 and now again approaching 2025, manufacturers are still producing semiconductors every 2 years that nearly double the number of transistors.
The linear regression model is much better at predicting the average than extreme values because it satisfies the condition to minimize \(\sum |y_i - A\cdot \text{year}[i]+B|^2\).
Wrapping up#
In conclusion, you have compared historical data for semiconductor
manufacturers to Moore’s law and created a linear regression model to
find the average number of transistors added to each microprocessor
every two years. Gordon Moore predicted the number of transistors would
double every two years from 1965 through 1975, but the average growth
has maintained a consistent increase of \(\times 1.98 \pm 0.01\) every two
years from 1971 through 2019. In 2015, Moore revised his prediction to
say Moore’s law should hold until 2025.
[2].
You can share these results as a zipped NumPy array file,
mooreslaw_regression.npz, or as another csv,
mooreslaw_regression.csv. The amazing progress in semiconductor
manufacturing has enabled new industries and computational power. This
analysis should give you a small insight into how incredible this growth
has been over the last half-century.
